Digestion and ligation to adaptors
PCR amplification with adaptor specific primers
Resulting band pattern
Suggestion of restriction enzymes pairs for AFLP-PCR
Introduction
Amplified Fragment Length Polymorphism PCR (AFLP-PCR) was originally described by Zabeau & Vos in 1993. The procedure of this technique is divided into three steps:
- Digestion of total cellular DNA with one or more restriction enzymes and ligation of restriction half-site specific adaptors to all restriction fragments.
- Selective amplification of some of these fragments with two PCR primers that have corresponding adaptor and restriction site specific sequences.
- Electrophoretic separation and amplicons on a gel matrix, followed by visualisation of the band pattern.
Choosing bacteria species
Select the bacterial genera you want to work with. A form will appear where we may select the species, and the restriction enzymes and selective nucleotides for the experiment.
We may also include the plasmids if available. For bacterial species with two chromosomes, both chromosomes will be used in the experiment. In some bacterial species, chromosomes are linear, and this fact has also been considered in the experiment.
Digestion and ligation to adaptors
After selection of genome, the information necessary to perform the experiment must be selected. The form has been partially reproduced below:
5'-
-3'
Select restriction enzymes from the lists (restriction enzyme 1 from RE1 list, and restriction enzyme 2 from RE2 list). They will be used to perform a complete theoretical DNA digestion of the genome. Selective nucleotides may also be introduced (see selective nucleotides in the form above: "AG" in the 5' end, and "GT" in the 3' end).
The restriction enzymes available in the form have the following characterictics:
- They all will cleave DNA within recognition sequence
- The recognized sequence is not ambiguous (no degenerated nucleotides)
- They will all yeild overhang ends (no blund ends)
As a result of DNA digestion, three types of DNA fragments will be produced:
- Fragments cleaved in both ends by the same restriction enzyme (RE1 or RE2)
- Fragments cleaved in 5' end by RE1 and by RE2 in 3' end
- Fragments cleaved in 3' end by RE1 and by RE2 in 5' end
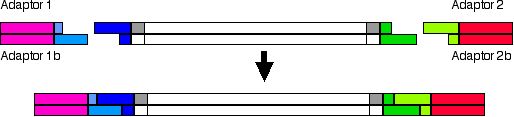
Ligation of adaptors in AFLP-PCR experiments
The sequence of these adaptors are partially defined in the response page. Some nucleotides of the adaptors are defined by the recognition sequence of the restriction enzymes (in pale blue and pale green). The sequence of adaptors which does not match the endonuclease recognition sequence (in magenta and red) must be designed to avoid recognition of genomic DNA of the species used in the experiment and to prevent aberrant results.
If the restriction enzymes used to cut the genomic DNA are not heat labile, or restriction and ligation are performed simultaneously, the adaptor sequence must not regenerate the original recognition sequence. To avoid this regeneration they must be used adaptors mustbe used that produce a base change in the recognition sequence. An example is shown below:
//-------GAATTC------//------TTAA------// //-------CTTAAG------//------AATT------// EcoRI MseI |
DNA sequence with EcoRI and MseI recognition sequences |
//-------G AATTC------//------T TAA------// //-------CTTAA G------//------AAT T------// |
DNA restriction |
NNNNNNA AATTC------//------T TACnnnnnn nnnnnnTTTAA G------//------AAT GNNNNNN NNNNNNAAATTC------//------TTACnnnnnn nnnnnnTTTAAG------//------AATGNNNNNN |
Addition of adaptors. As nucleotides in red are different to the original ones (blue), restriction sites are not reconstructed. |
In the results page of theoretical AFLP-PCR partial sequence of adaptors allow the reconstruction of th original recognition sequence, which is a valid option only when heat labile endonucleases are used, and restriction and ligation are performed separately.
PCR amplification with adaptor specific primers
In AFLP-PCR experiments, the primers must be designed to allow PCR amplification of the fragments cleaved by RE1 in 5' end and RE2 in 3' end, so that they will be complementary to sequence defined by adaptors and sequence recognised by restriction enzymes. The amplification will be performed as shown in the picture:
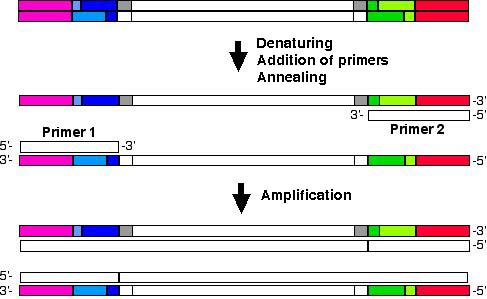
AFLP-PCR with primers matching adaptors and recognised restriction enzymes sequences.
Partial sequence of primers necessary to perform AFLP-PCR are suggested in the response page.
When experiments are perfformed as shown in this page, several combinations of restriction enzymes used in AFLP-PCR will yeild a large number of amplicons, so that interpretation will be very difficult. In fact, AFLP-PCR experiments performed by this service will not show bands when their number is above 50.
In order to avoid getting a large number of bands, in clasical AFLP-PCR, two consecutive PCR amplifications are performed. The first one is performed with primers which match only adaptors and sequences recognized by restriction enzymes (as in the experiment above). The resulting amplicons are used in a second PCR with slightly longer primers: those primers will match adaptors, restriction enzyme recognized sequence and they will contain one or more nucleotides in the 3' end (selective nucleotides), so that the number of amplicons yeilded in this second PCR are fewer (as shown in the picture below). This way, thenumber of bands will be lower and interpretation of band pattern easier.

AFLP-PCR with primers matching adaptors, restriction enzymes recognised sequences and containning aditional selective bases in the 3' end. Usage of selective primers will reduce the number of bands amplified.
In silico AFLP-PCR experiment allows the calculation of band pattern yeilded when selective bases are added to primers. The selective bases must be introduced in the form as shown above.
Resulting bands pattern
After clicking "Amplify" buttom, the following data will be available:
- Selected restriction enzyme, all isoschizomers, and
recognition sequences.
A link will allow to find vendors. - Partial sequence of adaptors and primers.
- Position and length of bands within genome.
- Band pattern after electrophoresis in agarose gel.
- A link will allow the following data to obtained
- DNA sequence of each amplicon
- Open Reading Frames (ORF) to which the amplicon belongs
- Links to NCBI server to get genomic maps
REFERENCES
Zabeau, M and P. Vos. 1993. Selective restriction fragment amplification: a general method for DNA fingerprinting. European Pattent Office, publication 0 534 858 A1, bulletin 93/13.
2003-2015@ University of the Basque Country. All
rights reserved.
