In silico Double Digestion
Fingerprinting (DDF)
|
EcoRI-AflI fragments |
As it can be seen in the example, the AflI overhang ends are different in type A and B fragments, and in type C and D fragments. Type A and D fragments and type B and C are identical.
To amplify all fragments we will need adaptors for EcoRI end and for the two types of AflI ends (AflI adapters A and B).
Adapters required to amplified all EcoRI-AflI and AflI-EcoRI fragments |
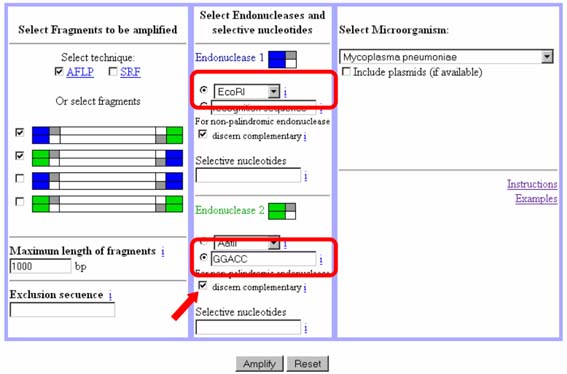
In this experiment, we have selected the checkbox indicated by the arrow ("For non-palindromic endonuclease, discern complementary"). By selecting this checkbox we are indicating to the program that fragments from EcoRI to AflI must be computed (and AflI to EcoRI) where the recognition sequence for AflI is GGACC, but not the complementary (GGTCC). So the recognition sequence indicated is discerned from its complementary. As a consequence, the program will only compute fragment types A and D (and fragment B and C will not be computed).
The use of non-palindromic endonucleases in AFLP-PCR experiments may be considered as a strategy to reduce the number of fragments amplified. The effect of this strategy will be similar to the ue of selective nucleotides (reduction of number of amplified fragments). In this experiment with M genitalium, from a total of 152 cEoRI-AflI fragments (fragments types A, B, C and D), only 94 are amplified (fragments types A and D).
Two non-palindromic endonucleases may be used in the experiment, and selective nucleotides may also be included in the experiment.
2005-2015@ University of the Basque Country. All
rights reserved.